GenRA Manual: Uncertainty Assessment and Conclusions
Predictions made by GenRA can be exported by clicking on the “Download” button (Figure 25). The file can be downloaded in either CSV or Excel (xlsx) format.
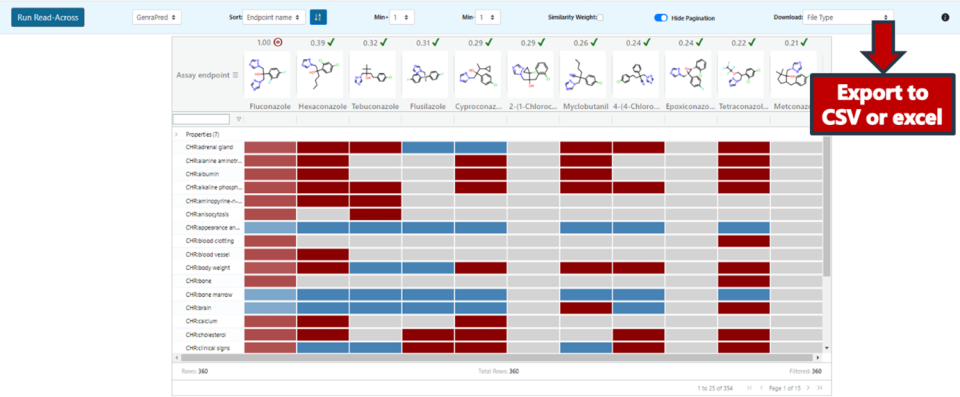
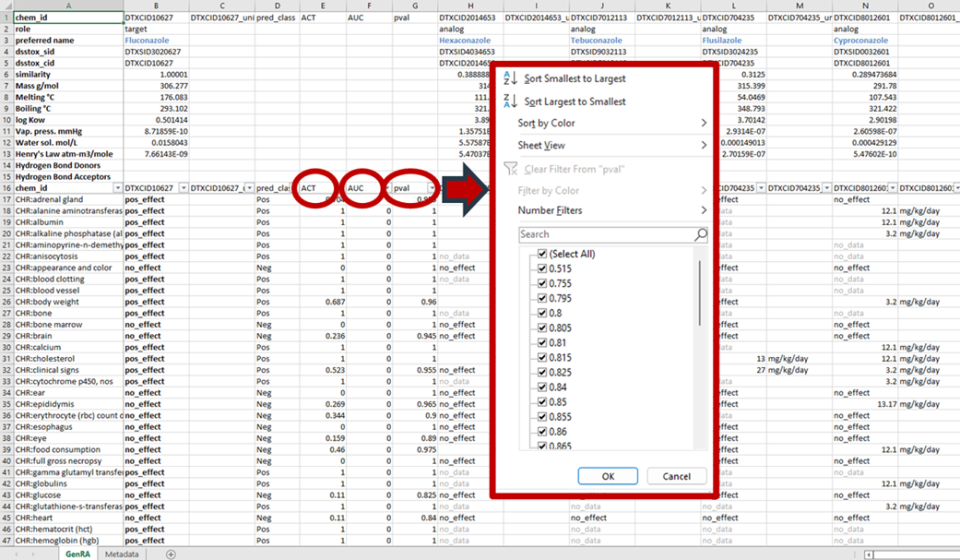
The AUC performance measure and the p-value associated with each prediction are provided, when relevant. This can be examined when the predictions generated are downloaded as a CSV file or Excel (xlsx) file (Figure 26). Both the exported file and in-app predictions can be sorted by p-values (high p-values ⇒ lack of confidence in the predictions). The web app allows for various other sorting methods as well, such as prioritizing positive or negative predictions. The actual experimental data for the source analogues is also reflected in the exported file.
Other Options
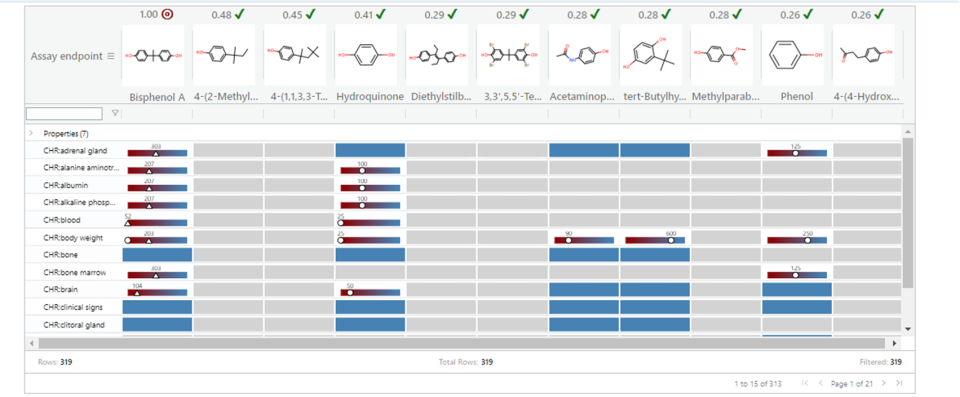
To generate potency predictions, the Tox Fingerprint Dosage needs to be selected in panel 3. This will change the view in panel 4. Instead of cells of red and blue boxes to reflect presence and absence of toxicity, the potency of a substance is shown and expressed in units of mg/kg bw. Each cell is scaled by the range of potencies (expressed in terms of a log10 molar basis) across a specific study type. This is intended to provide a visual cue of how potent a given source analogue is relative to the range of potencies observed for that study type. To make GenRA predictions, GenraPy is the automatic default which upon clicking Run Read-Across, updates the target substance column with predictions. Predicted values are shown on the transformed scale and represented as triangles with the specific value shown in units of mg/kg bw. If a study-toxicity value was already known, both the experimental value (denoted by a circle) and the predicted value (denoted by a triangle) are shown on the same scale. The hover over for the target outcomes provides the transformed and untransformed actual and predicted values. Figure 27 shows the updated panel 4 to illustrate how the actual and predicted values are presented in the data matrix.
Questions/Further information
For further information: contact us at [email protected]. This email is provided as the Contact link on the main application page.
Next chapter: Other Fingerprint Types
